![]()
BLUE MOLD
![]()
BLUE MOLD
Peronospora tabacina Adam is
the causal agent of blue mold (downy mildew) of tobacco and belongs to
the Oomycetes, a diverse group of fungus-like organisms that cause a wide
range of destructive and economically important diseases on plants.
 |
 |
Abundant sporulation results in a bluish-grey,
velvet-like growth on the host leaf
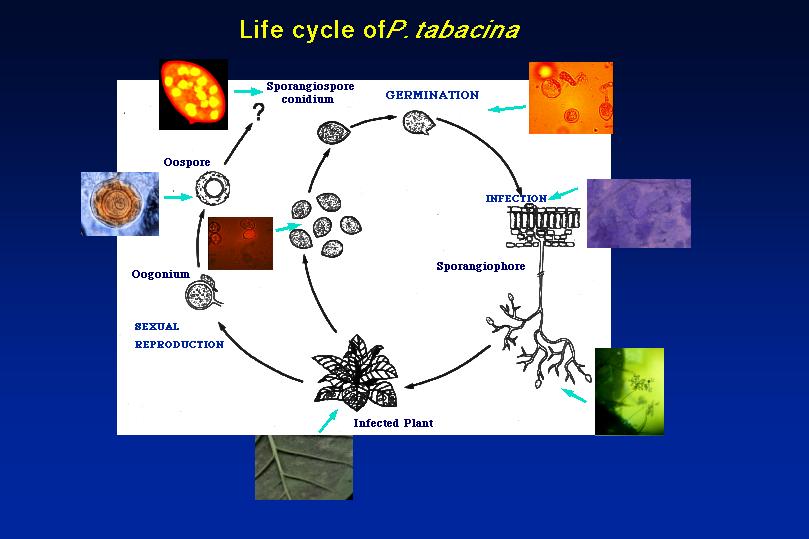
Life cycle of Peronospora tabacina
Very little is known about genetics
or molecular biology of P.tabacina. This is largely due the difficulties
in isolating DNA from obligate parasites, which require living host tissues
for growth and which cannot be isolated in pure culture. To generate molecular
markers for this organisms, sporangiospores were collected from infected
tobacco leaves as shown below.
Peronospora tabacina single
spore for DNA isolation
Use of molecular markers for epidemiological and population studies of Peronospora tabacina
We used a collection of random genomic DNA fragments from Peronospora tabacina to survey for restriction Fragment Lengh Polymorphisms(RFLPs) in DNA from a representative collection of isolates from Nicotiana tabacum in the Unites States. Also included in the study were isolates from the wild tobacco species N. rapanda. In our preliminary survey using DNA from 10 pathogen isolates, no polymorphisms were detected at 6 single-copy DNA loci, using 22 probe-enzyme combinations. Moderately-repetitive and highly-repetitive regions of the genome were also remarkably similar between isolates, with only 6 out of 15 different probes identifying genetic differences. Some of the polymorphic probes were then used to analyze a larger collection of isolates. This resulted in the identification of very few additional polymorphisms. Together, these results indicate that the US population of P. tabacina is genetically very homogeneous. Two of the RFLP markers gave hybridization patterns that were consistent with P. tabacina being diploid. Frequencies of alleles at these loci and linkage desequilibrium between different marker loci indicated that genetic recombination does not occur frequently in the US population of P. tabacina. DNA polymorphisms that were identified in this study enabled us to differentiate the pathogen population into at least 10 haplotypes. One isolate was analyzed in detail and was shown to be genetically stable through several rounds of single spore isolation and pathogenic growth. Despite their small numbers, the markers identified in this study were quite useful for differentiating among isolates, enabling resolution of at least ten genetically different forms of P. tabacina in the collection of isolates analyzed. In this regard, these markers promise to be valuable for studying numerous aspects of P. tabacina population biology.Ultimately, this will lead to improved strategies for management of blue mold
![]()
Blue mold warning system
Use od Molecular Markers for Population Studies of the Blue Mold Pathogen, Peronospora tabacina. 39th Tobacco Worker’s Conference. Colonial Williamsburg, Virginia. January 10-13.
Sukno, S, Taylor, A., Nesmith, W. and Farman, M. 2000.
ABSTRACT
Peronospora tabacina Adam is an oomycete that casues blue mold (downy mildew) a common and potentially severe desease of tabacco in kentucky and other states. This desease has caused significant crop losses in the United States every year since 1979. It has been hypothesized that the 1979 epidemic was due to a race change in the pathogen population (Lucas, 1980) but in the absence of methods to reliably characterize the pathogen population, this hypothesis was untestable. Molecular markers based on Restriction Fragment Length Polymorphisms (RFLPs) were developed using low copy, middle and highly repetitive DNA probes. These markers will enable identification and tracking of Peronospora tabacinapopulations. Results obtained using these markers to characterize US Blue Mold Populations will be described.
RFLP markers for population studies of the blue mold pathogen, Peronospora tabacina. Phytopathology 90:S75.
S. SUKNO, A. Taylor and M. Farman. 2000.
ABSTRACT
Blue mold of tobacco, caused by the oomycete, Peronospora tabacina, is an economically important disease. To examine the genetics and population biology of this organism RFLP markers were developed using low copy, middle copy and highly repetitive DNA probes isolated from partial genomic DNA libraries. Preliminary analysis of 9 blue mold strains indicates that most isolates are geneticallysimilar. However, DNA from 2 isolates failed to hybridize to several probes. These preliminary data suggest that although there is more than one genetic form of blue mold in the US, a single clone predominates. This hypothesis will be tested by RFLP analysis of a more comprehensive collection of isolates. Some DNA probes produced inconsistent and/or irreproducible hybridization patterns in different DNA preps of a single isolate. Based on DNA sequence analysis, we conclude that these probes were derived from bacteria. These could be either associated with P. tabacina infections or they might be endosymbionts. Regardless, the unavoidable presence of contaminating bacterial DNA indicates that results from PCR-based analyses of P. tabacina maybe unreliable.
Genetic uniformity among US isolates of Peronospora tabacina, the Tobacco Blue mold pathogen
S. SUKNO, A. Taylor and M. Farman. 2000.
Journal: Phytophatology. Submitted
ABSTRACT
We used a collection of random genomic DNA fragments from Peronospora tabacina to survey for restriction Fragment Lengh Polymorphisms(RFLPs) in DNA from a representative collection of isolates from Nicotiana tabacum in the Unites States. Also included in the study were isolates from the wild tobacco species N. rapanda. In a preliminary survey using DNA from 10 pathogen isolates, no polymorphisms were detected at 6 single-copy DNA loci, using 22 probe-enzyme combinations. Moderately-repetitive and highly-repetitive regions of the genome were also remarkably similar between isolates, with only 6 out of 15 different probes identifying genetic differences. Some of the polymorphic probes were then used to analyze a larger collection of isolates. This resulted in the identification of very few additional polymorphisms. Together, these results indicate that the US population of P. tabacina is genetically very homogeneous. Two of the RFLP markers gave hybridization patterns that were consistent with P. tabacina being diploid. Frequencies of alleles at these loci and linkage desequilibrium between different marker loci indicated that genetic recombination does not occur frequently in the US population of P. tabacina. DNA polymorphisms that were identified in this study enabled us to differentiate the pathogen population into at least 10 haplotypes. One isolate was analyzed in detail and was shown to be genetically stable through several rounds of single spore isolation and pathogenic growth.
Development and characterization of RFLP probes for Peronospora tabacina, an obligate parasite causing blue mold of tobacco.
S. SUKNO, A. Taylor and M. Farman. 2001.
Journal: Phytophatology. Submitted
ABSTRACT
Peronospora tabacina is an obligate parasitic oomycete that causes blue mold, a devastating disease of tobacco. To generate molecular markers for this organism, sporangiospores were collected from infected tobacco leaves, DNA was extracted and cloned in a plasmid vector. The resulting clones were then used to probe DNA from a collection of P. tabacina isolates to survey for polymorphisms. Most probes gave unexpected hybridization patters with signal intensities that varied significantly from one DNA sample to another. In addition, signals varied between different DNA preparations of the same isolate. These results gave an indication that some probes were derived from contaminating organisms present in the spore suspensions. Therefore, we characterized the inserts of several recombinant plasmids to determine their origins. Sequence analysis revealed that several of the inserts encoded peptides with similarity to bacterial proteins, suggesting that they were derived from bacterial contaminants. Of the remaining clones, five exhibited similarity to retroelements, one resembled eukaryotic helicase genes, and nine had no similarity to sequences in the databases. These were postulated to be true P. tabacina DNA clones. Verification of the origin of each probe was achieved by filtering a spore suspension, extracting DNA from the retentate and filtrate and probing Southern blots of these DNA samples. Theses experiments confirmed the probe origins predicted by the sequencing analysis. This study resulted in the generation of 28 confirmed P. tabacina probes, thereby providing reliable molecular markers for population studies of the blue mold organism.
Analysis of genetic variation in Peronospora tabacina using RFLPs. XXI Fungal Genetis Conference, Asilomar CA.
S. SUKNO and M. Farman. 2001.
ABSTRACT
Peronospora tabacina Adam is the causal agent of blue mold of tobacco and belongs to the Oomycetes, a diverse group of fungus-like organisms that cause a wide range of destructive and economically important diseases on plants. The inability to identify and track specific P. tabacina populations hamper efforts to control blue mold. Such information is vital to the successful implementation of durable disease management strategies in US. To examine the genetics and population biology of this obligate biotrophic parasite, three Pst-I genomic DNA libraries were constructed from DNA of three isolates that originated from Kentucky, USA. In preparation for a broader population study, 10 strains representing populations from Kentucky, Florida, Texas, Georgia, Pennsylvania, and Connecticut, were selected for an initial survey of RFLPs markers. Pst-I and Dra-I digested DNA were hybridized to 10 probes. Preliminary analysis indicates that there is a low level of genetic variation among the US populations. Two polymorphic probes were identified and seven different haplotypes could be differentiated among 10 isolates. Experiments are currently under way to evaluate the somatic stability of single spore lineages of one isolate and variability among individuals in a population. Results of the studies will be presented.
Copyright © 2002 S Sukno